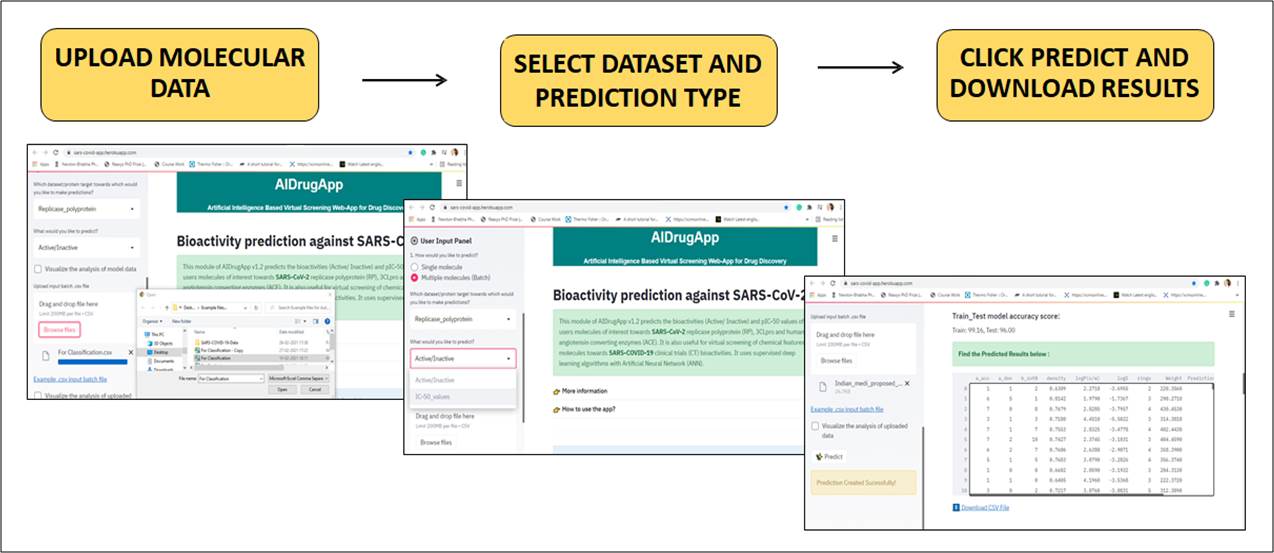
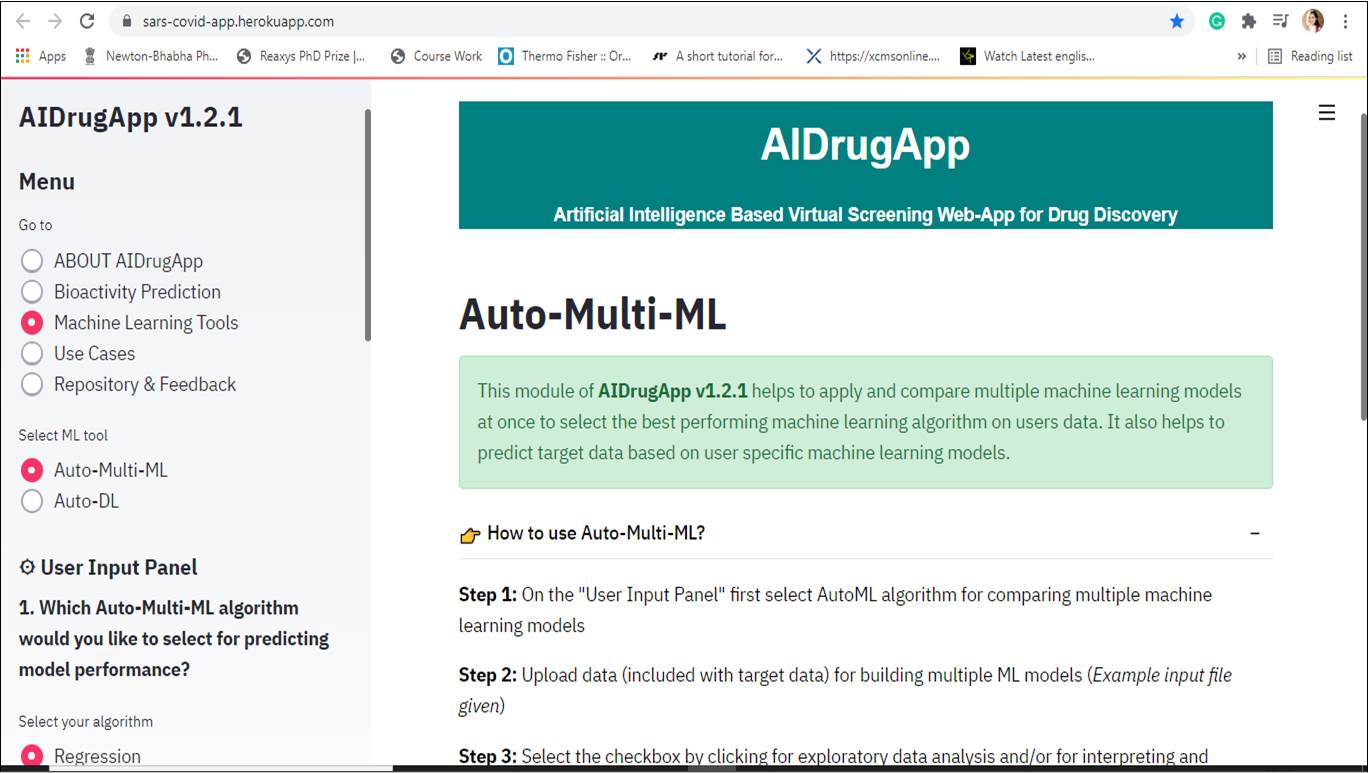
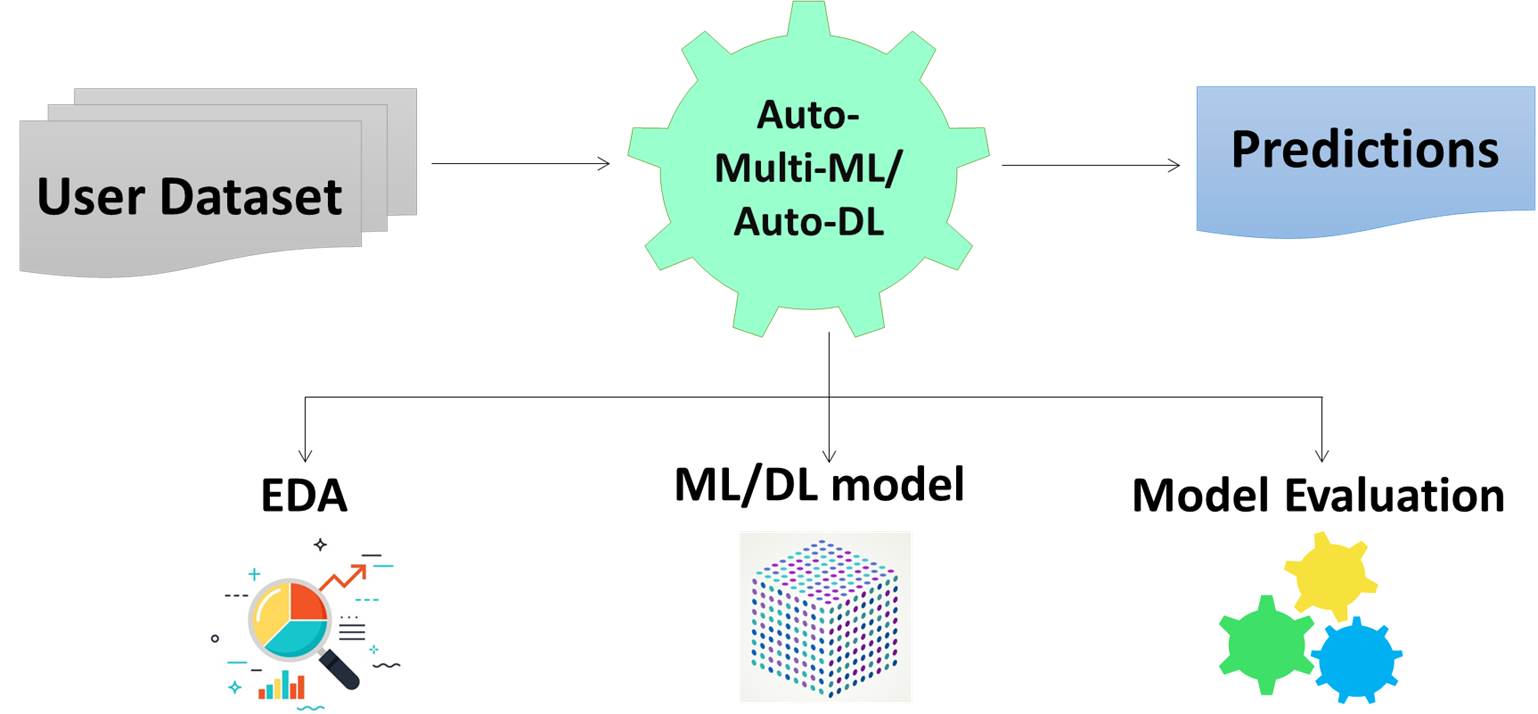
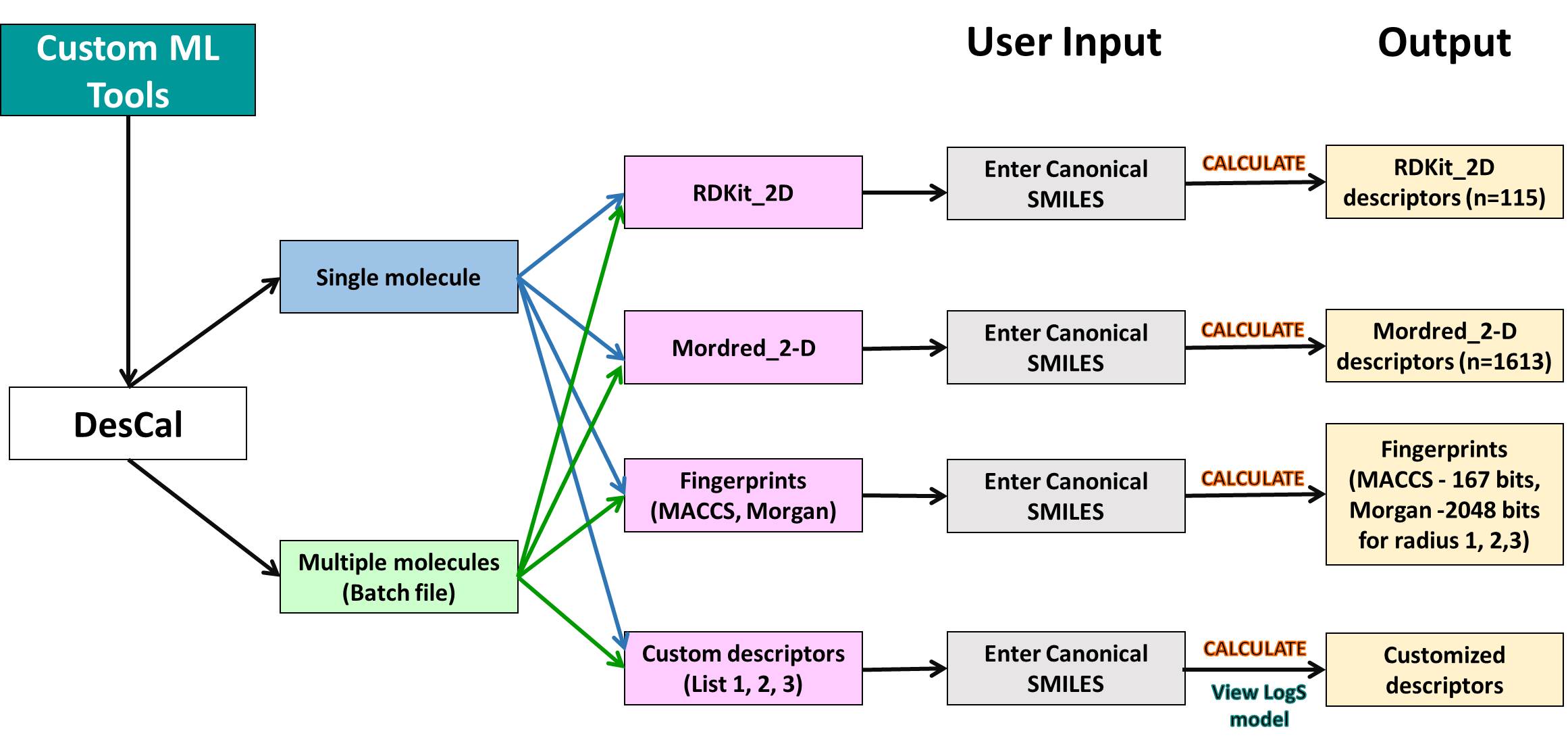
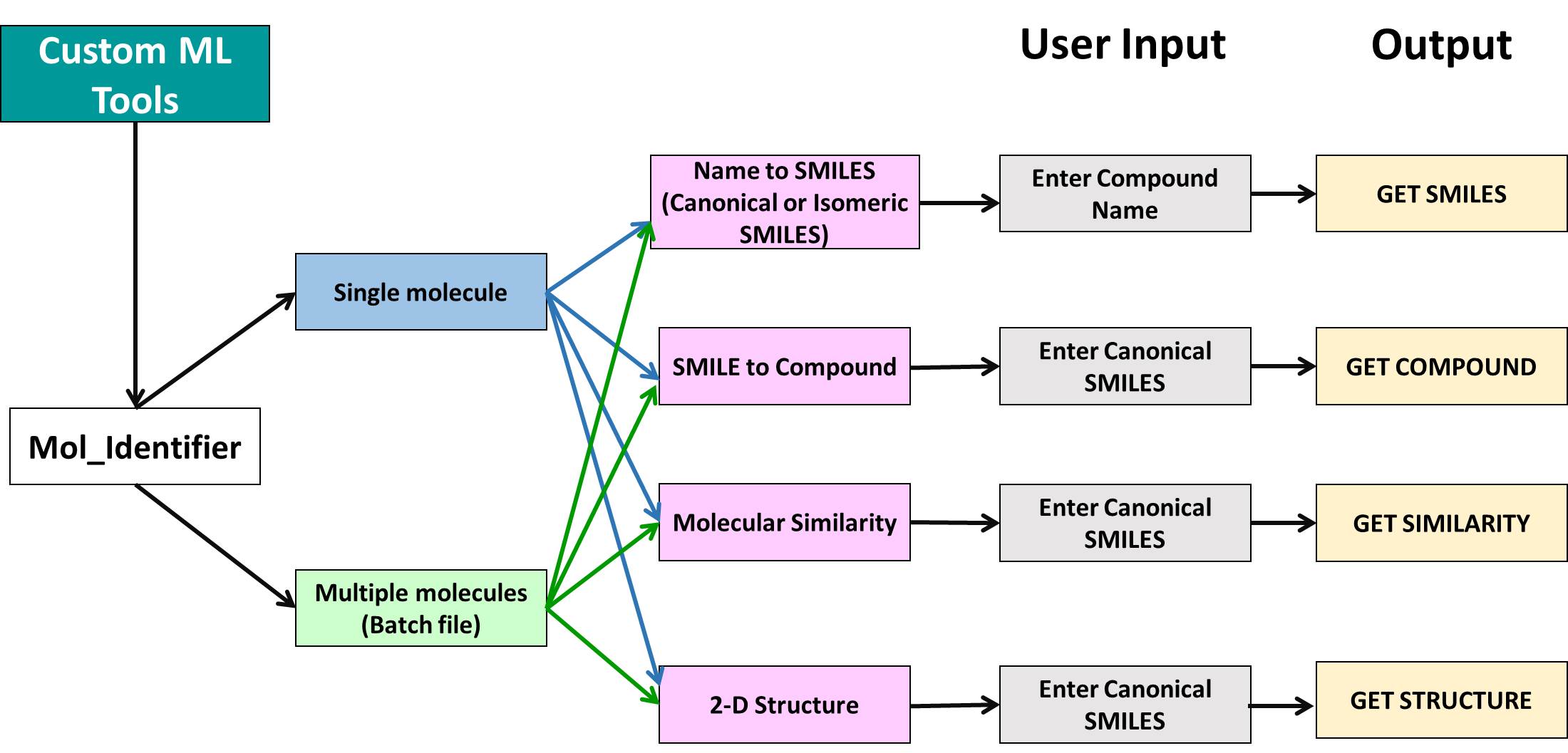
Bioactivity prediction against SARS-CoV-2
Bioactivity prediction is one the module of AIDrugApp v1.2.5 that helps to predict the bioactivities (Active/ Inactive) and pIC-50 values of users molecules of interest towards SARS-CoV-2 replicase polyprotein (RP), 3CLpro and human angiotensin converting enzymes (ACE). It is also useful for virtual screening of chemical features of molecules towards SARS-COVID-19 clinical trials (CT) bioactivities. It uses supervised deep learning algorithms with Artificial Neural Network (ANN).